Feature scaling and transformation in machine learning
In this tutorial we will learn how to scale or transform the features using different techniques.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
diabetes=pd.read_csv('diabetes_cleaned.csv')
features_df= diabetes.drop('Outcome',axis = 1)
target_df=diabetes['Outcome']
features_df.describe()
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | |
|---|---|---|---|---|---|---|---|---|
| count | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 |
| mean | 3.845052 | 121.539062 | 72.405184 | 29.108073 | 152.222767 | 32.307682 | 0.471876 | 33.240885 |
| std | 3.369578 | 30.490660 | 12.096346 | 8.791221 | 97.387162 | 6.986674 | 0.331329 | 11.760232 |
| min | 0.000000 | 44.000000 | 24.000000 | 7.000000 | -17.757186 | 18.200000 | 0.078000 | 21.000000 |
| 25% | 1.000000 | 99.000000 | 64.000000 | 25.000000 | 89.647494 | 27.300000 | 0.243750 | 24.000000 |
| 50% | 3.000000 | 117.000000 | 72.202592 | 29.000000 | 130.000000 | 32.000000 | 0.372500 | 29.000000 |
| 75% | 6.000000 | 140.250000 | 80.000000 | 32.000000 | 188.448695 | 36.600000 | 0.626250 | 41.000000 |
| max | 17.000000 | 199.000000 | 122.000000 | 99.000000 | 846.000000 | 67.100000 | 2.420000 | 81.000000 |
As you can see range of columns are very high for e.g. Age range is 21-81. ML models always give you better result when all of the features are in same range. So let’s do that using various techniques.
Feature Scaling and Standardization
When all features are in different range then we change the range of those features to a specific scale ,this method is called feature scaling.
- Normalization and Standardization are two specific Feature Scaling methods.
Min Max Scaler
from sklearn.preprocessing import MinMaxScaler
scaler=MinMaxScaler(feature_range=(0,1))
rescaled_features=scaler.fit_transform(features_df)
rescaled_features
array([[0.35294118, 0.67096774, 0.48979592, ..., 0.31492843, 0.23441503,
0.48333333],
[0.05882353, 0.26451613, 0.42857143, ..., 0.17177914, 0.11656704,
0.16666667],
[0.47058824, 0.89677419, 0.40816327, ..., 0.10429448, 0.25362938,
0.18333333],
...,
[0.29411765, 0.49677419, 0.48979592, ..., 0.16359918, 0.07130658,
0.15 ],
[0.05882353, 0.52903226, 0.36734694, ..., 0.24335378, 0.11571307,
0.43333333],
[0.05882353, 0.31612903, 0.46938776, ..., 0.24948875, 0.10119556,
0.03333333]])
rescaled_diabetes = pd.DataFrame(rescaled_features, columns=features_df.columns)
rescaled_diabetes
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0.352941 | 0.670968 | 0.489796 | 0.304348 | 0.274029 | 0.314928 | 0.234415 | 0.483333 |
| 1 | 0.058824 | 0.264516 | 0.428571 | 0.239130 | 0.101819 | 0.171779 | 0.116567 | 0.166667 |
| 2 | 0.470588 | 0.896774 | 0.408163 | 0.239130 | 0.333110 | 0.104294 | 0.253629 | 0.183333 |
| 3 | 0.058824 | 0.290323 | 0.428571 | 0.173913 | 0.129385 | 0.202454 | 0.038002 | 0.000000 |
| 4 | 0.000000 | 0.600000 | 0.163265 | 0.304348 | 0.215057 | 0.509202 | 0.943638 | 0.200000 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 763 | 0.588235 | 0.367742 | 0.530612 | 0.445652 | 0.228950 | 0.300613 | 0.039710 | 0.700000 |
| 764 | 0.117647 | 0.503226 | 0.469388 | 0.217391 | 0.204424 | 0.380368 | 0.111870 | 0.100000 |
| 765 | 0.294118 | 0.496774 | 0.489796 | 0.173913 | 0.150224 | 0.163599 | 0.071307 | 0.150000 |
| 766 | 0.058824 | 0.529032 | 0.367347 | 0.239130 | 0.221796 | 0.243354 | 0.115713 | 0.433333 |
| 767 | 0.058824 | 0.316129 | 0.469388 | 0.260870 | 0.121509 | 0.249489 | 0.101196 | 0.033333 |
768 rows × 8 columns
rescaled_diabetes.boxplot(figsize=(12,10),rot=45)
<matplotlib.axes._subplots.AxesSubplot at 0x164058b0>
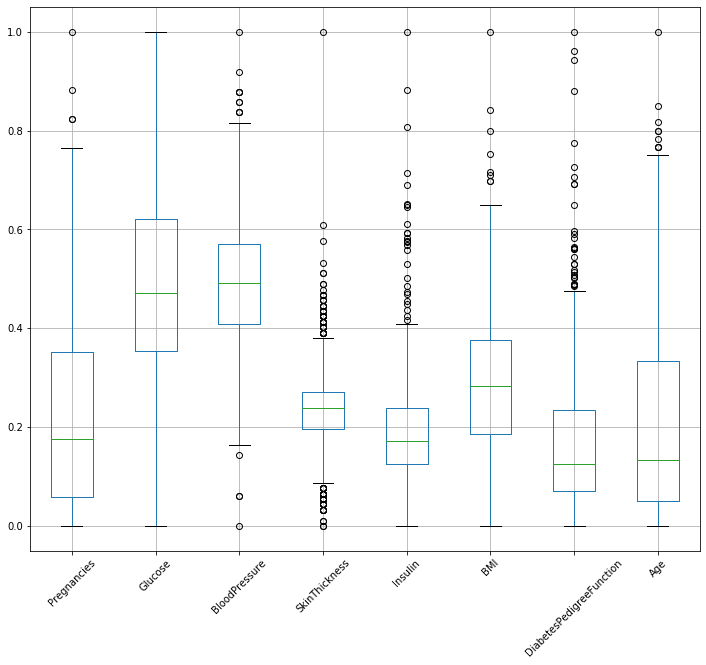
You can see all the data column’s range is between 0-1 now.
But here is one Catch!! MinMaxScaler is very sensitive to your data so make sure your whole prediction should not be hampered , like in this case if you see it has change age’s min to 0 but that is not the one we are looking for.
Standardization
Standardization is applied on feature wise , we calculate the mean of each feature then subtract each feature value from the mean and then divide it with the standard deivation Standardization centers mean of all our numeric features at 0 and expresses each value of the feature by the multiples of the std dev.
This is usually preferred because it is less outlier sensitive.
from sklearn.preprocessing import StandardScaler
scaler=StandardScaler()
standardized_features=scaler.fit_transform(features_df)
standardized_diabetes = pd.DataFrame(standardized_features, columns=features_df.columns)
standardized_diabetes
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0.639947 | 0.868403 | -0.033518 | 0.670643 | 0.685496 | 0.185089 | 0.468492 | 1.425995 |
| 1 | -0.844885 | -1.199150 | -0.529859 | -0.012301 | -0.842893 | -0.817471 | -0.365061 | -0.190672 |
| 2 | 1.233880 | 2.017044 | -0.695306 | -0.012301 | 1.209840 | -1.290106 | 0.604397 | -0.105584 |
| 3 | -0.844885 | -1.067877 | -0.529859 | -0.695245 | -0.598238 | -0.602636 | -0.920763 | -1.041549 |
| 4 | -1.141852 | 0.507402 | -2.680669 | 0.670643 | 0.162111 | 1.545707 | 5.484909 | -0.020496 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 763 | 1.827813 | -0.674057 | 0.297376 | 2.150354 | 0.285411 | 0.084833 | -0.908682 | 2.532136 |
| 764 | -0.547919 | 0.015127 | -0.198965 | -0.239949 | 0.067744 | 0.643403 | -0.398282 | -0.531023 |
| 765 | 0.342981 | -0.017691 | -0.033518 | -0.695245 | -0.413288 | -0.874760 | -0.685193 | -0.275760 |
| 766 | -0.844885 | 0.146400 | -1.026200 | -0.012301 | 0.221915 | -0.316191 | -0.371101 | 1.170732 |
| 767 | -0.844885 | -0.936604 | -0.198965 | 0.215347 | -0.668142 | -0.273224 | -0.473785 | -0.871374 |
768 rows × 8 columns
standardized_diabetes.boxplot(figsize=(12,10),rot=45)
<matplotlib.axes._subplots.AxesSubplot at 0x1679ec30>
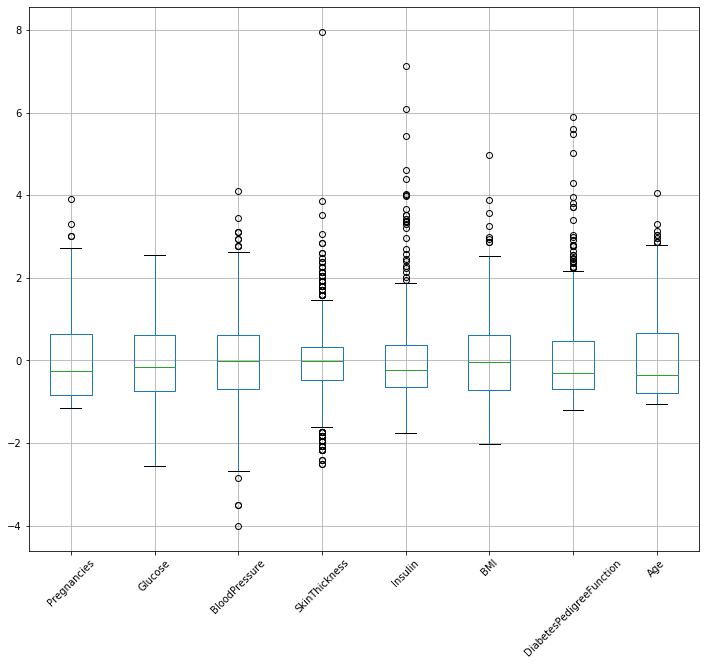
You can see that all features’ mean have been centered to zero and if any feature is not having many outliers then its median should not be far away from the mean.
Normalization
Normalization converts the feature vectors to their unit norm representations , there are different types of unit norms such as
- L1 Normalization
- L2 Normalization
- Max Normalization
This is not useful with data having outliers!
from sklearn.preprocessing import Normalizer
L1 Normailization
normalizer = Normalizer(norm='l1')
l1_normalized_features = normalizer.fit_transform(features_df)
l1_normalized_diabetes = pd.DataFrame(l1_normalized_features, columns=features_df.columns)
l1_normalized_diabetes.head()
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0.010635 | 0.262335 | 0.127622 | 0.062039 | 0.388074 | 0.059557 | 0.001111 | 0.088627 |
| 1 | 0.003235 | 0.274956 | 0.213495 | 0.093809 | 0.227047 | 0.086045 | 0.001135 | 0.100278 |
| 2 | 0.013116 | 0.300029 | 0.104928 | 0.047546 | 0.442615 | 0.038200 | 0.001102 | 0.052464 |
| 3 | 0.003103 | 0.276169 | 0.204799 | 0.071369 | 0.291684 | 0.087195 | 0.000518 | 0.065163 |
| 4 | 0.000000 | 0.298873 | 0.087262 | 0.076355 | 0.366502 | 0.094025 | 0.004991 | 0.071991 |
l1_normalized_diabetes.iloc[0]
Pregnancies 0.010635
Glucose 0.262335
BloodPressure 0.127622
SkinThickness 0.062039
Insulin 0.388074
BMI 0.059557
DiabetesPedigreeFunction 0.001111
Age 0.088627
Name: 0, dtype: float64
Every row in your dataset is a feature vector and normalization is a technique to convert those feature vector by their unit magnitude there are different types of unit magnitudes here we have converted using L1 unit magnitude.
In L1 normalization summation of absolute values of these normalized features is 1.
l1_normalized_diabetes.iloc[0].abs().sum()
1.0
L2 Normalization
In L2 normalization every feature vector or records in your dataset will be converted to their L2 unit magnitude and sum of the individual features’ square will be 1
normalizer = Normalizer(norm='l2')
l2_normalized_features = normalizer.fit_transform(features_df)
l2_normalized_diabetes = pd.DataFrame(l2_normalized_features, columns=features_df.columns)
l2_normalized_diabetes.head()
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0.021225 | 0.523547 | 0.254698 | 0.123812 | 0.774487 | 0.118859 | 0.002218 | 0.176874 |
| 1 | 0.007251 | 0.616359 | 0.478585 | 0.210287 | 0.508963 | 0.192884 | 0.002545 | 0.224790 |
| 2 | 0.023805 | 0.544535 | 0.190439 | 0.086292 | 0.803320 | 0.069332 | 0.002000 | 0.095219 |
| 3 | 0.006612 | 0.588467 | 0.436392 | 0.152076 | 0.621527 | 0.185797 | 0.001104 | 0.138852 |
| 4 | 0.000000 | 0.596386 | 0.174127 | 0.152361 | 0.731335 | 0.187622 | 0.009960 | 0.143655 |
l2_normalized_diabetes.iloc[0].pow(2).sum()
0.9999999999999997
Maximum Normalization
Now let’s talk about Maximum normalization here the maximum value of a feature vector is converted to 1 and other values of that feature vector will be converted in terms of this maximum.
normalizer = Normalizer(norm='max')
max_normalized_features = normalizer.fit_transform(features_df)
print(type(max_normalized_features))
max_normalized_diabetes = pd.DataFrame(max_normalized_features, columns=features_df.columns)
max_normalized_diabetes.head()
<class 'numpy.ndarray'>
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0.027405 | 0.675991 | 0.328861 | 0.159863 | 1.000000 | 0.153468 | 0.002864 | 0.228375 |
| 1 | 0.011765 | 1.000000 | 0.776471 | 0.341176 | 0.825756 | 0.312941 | 0.004129 | 0.364706 |
| 2 | 0.029633 | 0.677856 | 0.237064 | 0.107420 | 1.000000 | 0.086306 | 0.002489 | 0.118532 |
| 3 | 0.010638 | 0.946809 | 0.702128 | 0.244681 | 1.000000 | 0.298936 | 0.001777 | 0.223404 |
| 4 | 0.000000 | 0.815476 | 0.238095 | 0.208333 | 1.000000 | 0.256548 | 0.013619 | 0.196429 |
if you look at the above df you can see one feature in every record is transformed into 1 and other features are represented in terms of this max.
Binarizer
Now sometimes it may be required that we would want to discretize our numerical features there we can use binarizer. In binarizer we provide a threshold value for each feature and it will convert all values which is less than the threshold to zero and all values which is greater than the threshold to 1.
scaler=Binarizer(threshold=float((features_df[['Pregnancies']]).mean()))
binarized_features=scaler.fit_transform(features_df[['Pregnancies']])
from sklearn.preprocessing import Binarizer
for i in range(1,features_df.shape[1]):
scaler=Binarizer(threshold=float(features_df[features_df.columns[i]].mean())). \
fit(features_df[[features_df.columns[i]]])
new_binarized_feature = scaler.transform(features_df[[features_df.columns[i]]])
binarized_features = np.concatenate((binarized_features,new_binarized_feature),axis=1)
binarized_diabetes = pd.DataFrame(binarized_features, columns=features_df.columns)
binarized_diabetes.head(20)
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | |
|---|---|---|---|---|---|---|---|---|
| 0 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 |
| 3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 4 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 |
| 5 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 6 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 7 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 8 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 |
| 9 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 10 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 11 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 12 | 1.0 | 1.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 |
| 13 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 |
| 14 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 |
| 15 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 16 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.0 |
| 17 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 18 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 19 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 1.0 | 0.0 |
You can see all vectors have been represented using zero or 1 Now that we have transformed our data using different techniques let’s do some classification now.
Now lets build a classification model and see the differences:
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
def buildmodel(X,Y,test_frac):
x_train,x_test,y_train,y_test =train_test_split(X,Y,test_size=test_frac)
model =LogisticRegression(solver='liblinear').fit(x_train,y_train)
y_pred=model.predict(x_test)
print('Accuracy of the model is',accuracy_score(y_test,y_pred))
buildmodel(rescaled_diabetes,target_df,test_frac=.2)#using MinMaxScaler
Accuracy of the model is 0.7857142857142857
buildmodel(standardized_diabetes,target_df,test_frac=.2)#using StandardScaler
Accuracy of the model is 0.7662337662337663
buildmodel(l1_normalized_features,target_df,test_frac=.2)
Accuracy of the model is 0.6103896103896104
buildmodel(l2_normalized_features,target_df,test_frac=.2)
Accuracy of the model is 0.6623376623376623
buildmodel(max_normalized_features,target_df,test_frac=.2)
Accuracy of the model is 0.7337662337662337
buildmodel(binarized_features,target_df,test_frac=.2)
Accuracy of the model is 0.6753246753246753
You can get the notebook used in this tutorial here and dataset used here
Thanks for reading!






Leave a comment